DNA Repair Pathways and Cancer
Human cancers exhibit genomic instability and an increased mutation rate due to underlying defects in DNA repair. Cancer cells are often defective in one of six major DNA repair pathways, namely: mismatch repair, base excision repair, nucleotide excision repair, homologous recombination, nonhomologous endjoining and translesion synthesis. The specific DNA repair pathway affected is predictive of the kinds of mutations, the tumor drug sensitivity, and the treatment outcome. The study of rare inherited DNA repair disorders, such as Fanconi anemia, has yielded new insights to drug sensitivity and treatment of sporadic cancers, such as breast or ovarian epithelial tumors, in the general population. The Fanconi anemia pathway is an example of how DNA repair pathways can be deregulated in cancer cells and how biomarkers of the integrity of these pathways could be useful as a guide to cancer management and may be used in the development of novel therapeutic agents.
The Fanconi Anemia/BRCA Pathway Fanconi Anemia (FA) is an autosomal recessive or X-linked recessive cancer susceptibility syndrome characterized by multiple congenital abnormalities, progressive bone marrow failure, and cellular hypersensitivity to DNA crosslinking agents, such as cisplatin and mitomycin C (MMC). FA patients are prone to developing acute myeloid leukemia as well as squamous cell carcinomas of the head and neck or gynecologic system.
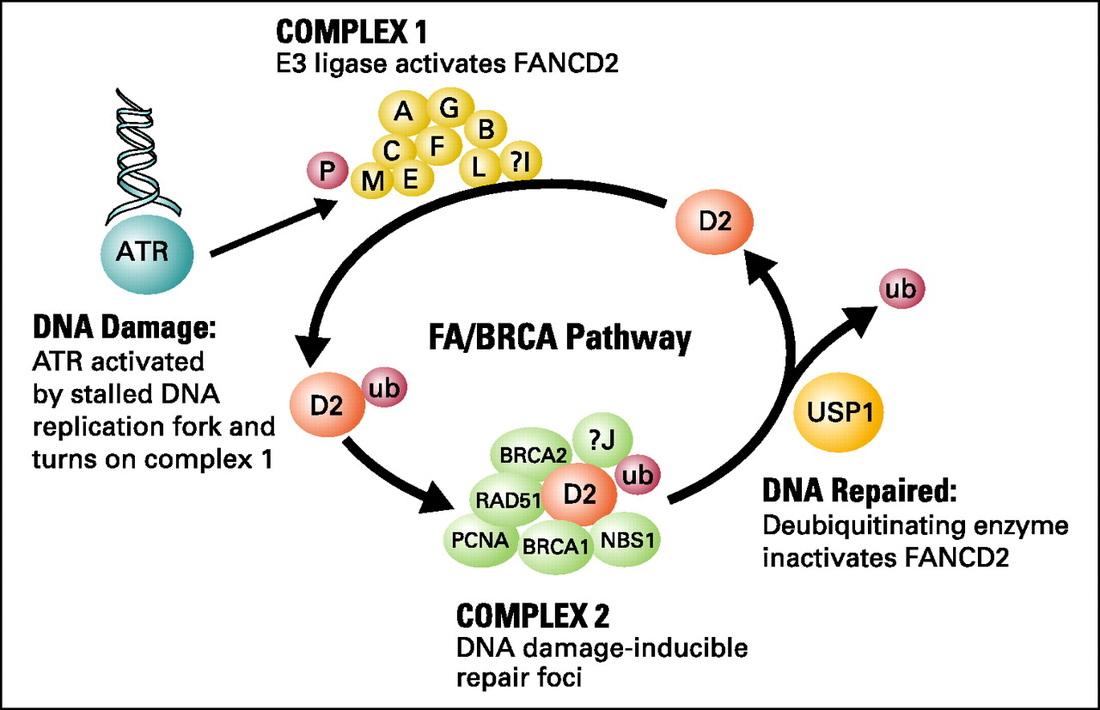
The study of FA cells has led to the elucidation of a DNA repair pathway for interstrand crosslinks. Clinically, this pathway is particularly important as many DNA crosslinking agents such as cisplatin, cyclophosphamide, mitomycin C or melphalan are used for cancer treatment. The FA defect results from biallelic mutation of any one of fifteen known FA genes (A, B, C, D1, D2, E, F, G, I, J, L, M, N, O, P). The proteins encoded by these FA genes cooperate in a common DNA repair pathway, referred to as the FA/BRCA pathway (Figure 1). In this pathway, eight of the FA proteins (A, B, C, E, F, G, L, M) are subunits of a nuclear E3 ubiquitin ligase, required for the monoubiquitination of the downstream D2 protein on lysine 561, which is a critical step for the function of the FA pathway. The FANCL subunit is the putative catalytic E3 ligase subunit of the complex. Monoubiquitinated FANCD2 (FANCD2-ub) interacts with FANCD1/BRCA2. The recently cloned FANCJ protein is a helicase which may work in concert with FANCD2-Ub and BRCA2 or may function independently of the FA pathway.
Recent research has suggested that the FA pathway may have a specific role in coordinating DNA repair pathways following DNA crosslinking damage. Consequently, disruption of any of the FA proteins leads to MMC hypersensitivity and chromosome instability.
The D’Andrea Laboratory is currently addressing several aspects of this novel DNA repair pathway including (1) the assembly, transport, and structure of the FA protein complex (2) the enzymatic monoubiquitination and deubiquitination of the D2 protein (3) the function of the chromatin-associated FA complex in cell cycle checkpoints and homologous recombination DNA repair (4) the identification of novel interacting proteins in these complexes.
Human cancers exhibit genomic instability and an increased mutation rate due to underlying defects in DNA repair. Cancer cells are often defective in one of six major DNA repair pathways, namely: mismatch repair, base excision repair, nucleotide excision repair, homologous recombination, nonhomologous endjoining and translesion synthesis. The specific DNA repair pathway affected is predictive of the kinds of mutations, the tumor drug sensitivity, and the treatment outcome. The study of rare inherited DNA repair disorders, such as Fanconi anemia, has yielded new insights to drug sensitivity and treatment of sporadic cancers, such as breast or ovarian epithelial tumors, in the general population. The Fanconi anemia pathway is an example of how DNA repair pathways can be deregulated in cancer cells and how biomarkers of the integrity of these pathways could be useful as a guide to cancer management and may be used in the development of novel therapeutic agents.
The Fanconi Anemia/BRCA Pathway Fanconi Anemia (FA) is an autosomal recessive or X-linked recessive cancer susceptibility syndrome characterized by multiple congenital abnormalities, progressive bone marrow failure, and cellular hypersensitivity to DNA crosslinking agents, such as cisplatin and mitomycin C (MMC). FA patients are prone to developing acute myeloid leukemia as well as squamous cell carcinomas of the head and neck or gynecologic system.
The study of FA cells has led to the elucidation of a DNA repair pathway for interstrand crosslinks. Clinically, this pathway is particularly important as many DNA crosslinking agents such as cisplatin, cyclophosphamide, mitomycin C or melphalan are used for cancer treatment. The FA defect results from biallelic mutation of any one of fifteen known FA genes (A, B, C, D1, D2, E, F, G, I, J, L, M, N, O, P). The proteins encoded by these FA genes cooperate in a common DNA repair pathway, referred to as the FA/BRCA pathway (Figure 1). In this pathway, eight of the FA proteins (A, B, C, E, F, G, L, M) are subunits of a nuclear E3 ubiquitin ligase, required for the monoubiquitination of the downstream D2 protein on lysine 561, which is a critical step for the function of the FA pathway. The FANCL subunit is the putative catalytic E3 ligase subunit of the complex. Monoubiquitinated FANCD2 (FANCD2-ub) interacts with FANCD1/BRCA2. The recently cloned FANCJ protein is a helicase which may work in concert with FANCD2-Ub and BRCA2 or may function independently of the FA pathway.
Recent research has suggested that the FA pathway may have a specific role in coordinating DNA repair pathways following DNA crosslinking damage. Consequently, disruption of any of the FA proteins leads to MMC hypersensitivity and chromosome instability.
The D’Andrea Laboratory is currently addressing several aspects of this novel DNA repair pathway including (1) the assembly, transport, and structure of the FA protein complex (2) the enzymatic monoubiquitination and deubiquitination of the D2 protein (3) the function of the chromatin-associated FA complex in cell cycle checkpoints and homologous recombination DNA repair (4) the identification of novel interacting proteins in these complexes.
DNA Repair Pathways and Cancer
Human cancers exhibit genomic instability and an increased mutation rate due to underlying defects in DNA repair. Cancer cells are often defective in one of six major DNA repair pathways, namely: mismatch repair, base excision repair, nucleotide excision repair, homologous recombination, nonhomologous endjoining and translesion synthesis. The specific DNA repair pathway affected is predictive of the kinds of mutations, the tumor drug sensitivity, and the treatment outcome. The study of rare inherited DNA repair disorders, such as Fanconi anemia, has yielded new insights to drug sensitivity and treatment of sporadic cancers, such as breast or ovarian epithelial tumors, in the general population. The Fanconi anemia pathway is an example of how DNA repair pathways can be deregulated in cancer cells and how biomarkers of the integrity of these pathways could be useful as a guide to cancer management and may be used in the development of novel therapeutic agents.
The Fanconi Anemia/BRCA Pathway Fanconi Anemia (FA) is an autosomal recessive or X-linked recessive cancer susceptibility syndrome characterized by multiple congenital abnormalities, progressive bone marrow failure, and cellular hypersensitivity to DNA crosslinking agents, such as cisplatin and mitomycin C (MMC). FA patients are prone to developing acute myeloid leukemia as well as squamous cell carcinomas of the head and neck or gynecologic system.
The study of FA cells has led to the elucidation of a DNA repair pathway for interstrand crosslinks. Clinically, this pathway is particularly important as many DNA crosslinking agents such as cisplatin, cyclophosphamide, mitomycin C or melphalan are used for cancer treatment. The FA defect results from biallelic mutation of any one of fifteen known FA genes (A, B, C, D1, D2, E, F, G, I, J, L, M, N, O, P). The proteins encoded by these FA genes cooperate in a common DNA repair pathway, referred to as the FA/BRCA pathway (Figure 1). In this pathway, eight of the FA proteins (A, B, C, E, F, G, L, M) are subunits of a nuclear E3 ubiquitin ligase, required for the monoubiquitination of the downstream D2 protein on lysine 561, which is a critical step for the function of the FA pathway. The FANCL subunit is the putative catalytic E3 ligase subunit of the complex. Monoubiquitinated FANCD2 (FANCD2-ub) interacts with FANCD1/BRCA2. The recently cloned FANCJ protein is a helicase which may work in concert with FANCD2-Ub and BRCA2 or may function independently of the FA pathway.
Recent research has suggested that the FA pathway may have a specific role in coordinating DNA repair pathways following DNA crosslinking damage. Consequently, disruption of any of the FA proteins leads to MMC hypersensitivity and chromosome instability.
The D’Andrea Laboratory is currently addressing several aspects of this novel DNA repair pathway including (1) the assembly, transport, and structure of the FA protein complex (2) the enzymatic monoubiquitination and deubiquitination of the D2 protein (3) the function of the chromatin-associated FA complex in cell cycle checkpoints and homologous recombination DNA repair (4) the identification of novel interacting proteins in these complexes.
Biomarkers of the FA Pathway in Cancer Treatment
Both hereditary cancer syndromes and sporadic cancers can arise from abnormalities in the FA pathway. Clinically, this may be important as these tumors are expected to be hypersensitive to DNA damaging therapeutic agents or strategies that inhibit alternative DNA repair pathways. In the case of the sporadic cancers, the patient’s normal cells, such as those in the bone marrow, should possess a functional pathway and would be predicted to be resistant to these targeted treatments. Assessment of the status of the FA pathway or other DNA repair pathways requires the use of diagnostic biomarkers.
Selection of Biomarkers of the FA Pathway
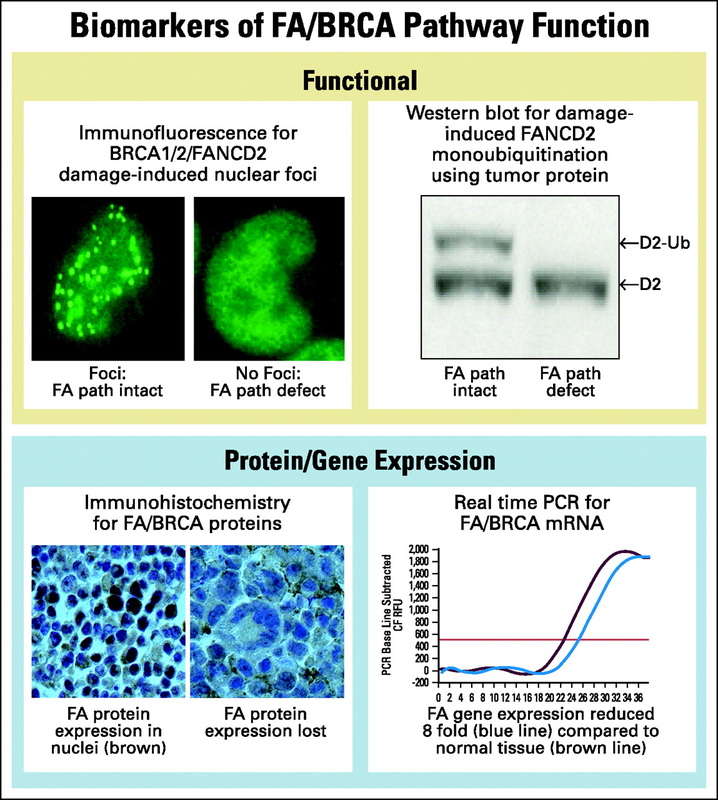
DNA repair biomarkers of the FA pathway can be divided into two major groups- functional biomarkers, that characterize the activity of a pathway following damage and expression biomarkers, that measure the availability of pathway components prior to damage (Figure 2).
Human cancers exhibit genomic instability and an increased mutation rate due to underlying defects in DNA repair. Cancer cells are often defective in one of six major DNA repair pathways, namely: mismatch repair, base excision repair, nucleotide excision repair, homologous recombination, nonhomologous endjoining and translesion synthesis. The specific DNA repair pathway affected is predictive of the kinds of mutations, the tumor drug sensitivity, and the treatment outcome. The study of rare inherited DNA repair disorders, such as Fanconi anemia, has yielded new insights to drug sensitivity and treatment of sporadic cancers, such as breast or ovarian epithelial tumors, in the general population. The Fanconi anemia pathway is an example of how DNA repair pathways can be deregulated in cancer cells and how biomarkers of the integrity of these pathways could be useful as a guide to cancer management and may be used in the development of novel therapeutic agents.
The Fanconi Anemia/BRCA Pathway Fanconi Anemia (FA) is an autosomal recessive or X-linked recessive cancer susceptibility syndrome characterized by multiple congenital abnormalities, progressive bone marrow failure, and cellular hypersensitivity to DNA crosslinking agents, such as cisplatin and mitomycin C (MMC). FA patients are prone to developing acute myeloid leukemia as well as squamous cell carcinomas of the head and neck or gynecologic system.
The study of FA cells has led to the elucidation of a DNA repair pathway for interstrand crosslinks. Clinically, this pathway is particularly important as many DNA crosslinking agents such as cisplatin, cyclophosphamide, mitomycin C or melphalan are used for cancer treatment. The FA defect results from biallelic mutation of any one of fifteen known FA genes (A, B, C, D1, D2, E, F, G, I, J, L, M, N, O, P). The proteins encoded by these FA genes cooperate in a common DNA repair pathway, referred to as the FA/BRCA pathway (Figure 1). In this pathway, eight of the FA proteins (A, B, C, E, F, G, L, M) are subunits of a nuclear E3 ubiquitin ligase, required for the monoubiquitination of the downstream D2 protein on lysine 561, which is a critical step for the function of the FA pathway. The FANCL subunit is the putative catalytic E3 ligase subunit of the complex. Monoubiquitinated FANCD2 (FANCD2-ub) interacts with FANCD1/BRCA2. The recently cloned FANCJ protein is a helicase which may work in concert with FANCD2-Ub and BRCA2 or may function independently of the FA pathway.
Recent research has suggested that the FA pathway may have a specific role in coordinating DNA repair pathways following DNA crosslinking damage. Consequently, disruption of any of the FA proteins leads to MMC hypersensitivity and chromosome instability.
The D’Andrea Laboratory is currently addressing several aspects of this novel DNA repair pathway including (1) the assembly, transport, and structure of the FA protein complex (2) the enzymatic monoubiquitination and deubiquitination of the D2 protein (3) the function of the chromatin-associated FA complex in cell cycle checkpoints and homologous recombination DNA repair (4) the identification of novel interacting proteins in these complexes.
Biomarkers of the FA Pathway in Cancer Treatment
Both hereditary cancer syndromes and sporadic cancers can arise from abnormalities in the FA pathway. Clinically, this may be important as these tumors are expected to be hypersensitive to DNA damaging therapeutic agents or strategies that inhibit alternative DNA repair pathways. In the case of the sporadic cancers, the patient’s normal cells, such as those in the bone marrow, should possess a functional pathway and would be predicted to be resistant to these targeted treatments. Assessment of the status of the FA pathway or other DNA repair pathways requires the use of diagnostic biomarkers.
Selection of Biomarkers of the FA Pathway
DNA repair biomarkers of the FA pathway can be divided into two major groups- functional biomarkers, that characterize the activity of a pathway following damage and expression biomarkers, that measure the availability of pathway components prior to damage (Figure 2).
Functional Biomarkers Markers
These are biomarkers that indicate an intact DNA repair pathway. These biomarkers have the advantage of giving a functional measure of a particular pathway and will detect repair defects due to epigenetic events or gene mutations. Moreover, they give a global measurement of a particular pathway’s function without needing to know the identities of all the components, some of which may remain unknown. They could also be used to differentiate between insignificant single nucleotide polymorphisms and functionally important point mutations in DNA repair pathway genes. Functional biomarkers can be applied to serial tumor samples from the same patient, at diagnosis and at the time of relapse. In this way, one can determine whether the tumor remains drug sensitive or has restored its DNA repair mechanisms. However they rely on tumor tissue having been exposed to some form of DNA damage in vivo or in vitro prior to the assay. Monoubuiquitination of FANCD2 is a biomarker for the integrity of the upstream part of the FA pathway. Abnormal DNA damage induced nuclear foci may identify disruption of the downstream events in the pathway, such as that observed in BRCA1 or BRCA2 deficient cells.
Biomarkers of Gene/Protein Expression
These biomarkers indicate the preexisting function of a DNA damage pathway prior to damage. Examples are real time PCR (rt-PCR) or immunohistochemistry to test for epigenetic silencing of critical DNA repair genes. Some studies have used a microarray approach to look for genetic expression profiles indicative of abnormal DNA repair gene function. Since some DNA repair genes, such as FANCF, undergo inactivation by methylation, the measurement of gene methylation, using the methylation-PCR assay, can also be applied as a biomarker assay. These approaches have the advantage of not requiring prior DNA damage and can be performed on fixed specimens. However, these assays provide only an indirect measurement of the functional capabilities of a DNA repair pathway. In addition, mutant genes can express normal levels of mRNA and mutant protein and would not be detected by this method.
These are biomarkers that indicate an intact DNA repair pathway. These biomarkers have the advantage of giving a functional measure of a particular pathway and will detect repair defects due to epigenetic events or gene mutations. Moreover, they give a global measurement of a particular pathway’s function without needing to know the identities of all the components, some of which may remain unknown. They could also be used to differentiate between insignificant single nucleotide polymorphisms and functionally important point mutations in DNA repair pathway genes. Functional biomarkers can be applied to serial tumor samples from the same patient, at diagnosis and at the time of relapse. In this way, one can determine whether the tumor remains drug sensitive or has restored its DNA repair mechanisms. However they rely on tumor tissue having been exposed to some form of DNA damage in vivo or in vitro prior to the assay. Monoubuiquitination of FANCD2 is a biomarker for the integrity of the upstream part of the FA pathway. Abnormal DNA damage induced nuclear foci may identify disruption of the downstream events in the pathway, such as that observed in BRCA1 or BRCA2 deficient cells.
Biomarkers of Gene/Protein Expression
These biomarkers indicate the preexisting function of a DNA damage pathway prior to damage. Examples are real time PCR (rt-PCR) or immunohistochemistry to test for epigenetic silencing of critical DNA repair genes. Some studies have used a microarray approach to look for genetic expression profiles indicative of abnormal DNA repair gene function. Since some DNA repair genes, such as FANCF, undergo inactivation by methylation, the measurement of gene methylation, using the methylation-PCR assay, can also be applied as a biomarker assay. These approaches have the advantage of not requiring prior DNA damage and can be performed on fixed specimens. However, these assays provide only an indirect measurement of the functional capabilities of a DNA repair pathway. In addition, mutant genes can express normal levels of mRNA and mutant protein and would not be detected by this method.